Revealing a hidden mechanism by which gut bacteria may influence cancer therapy
Key Takeaways:
- The specific interactions between human gut microbes and immune checkpoint inhibitor (ICI) cancer therapy have remained uncertain, with research coming to conflicting conclusions.
- High-throughput microbial isolation and cultivation using the Prospector® enabled the identification of six gut bacterial species that correlate with a therapy-responsive phenotype.
- Further study of secreted small molecules from bacterial cultures pointed to the active compound. This compound, when added to ICI therapy, shows promise in reducing tumor growth in mouse models.
- Additional details can be found in our on-demand webinar: “Small Microbes Make a Big Impact in Cancer Immunotherapy.
The human gut microbiome is known to influence health in a multitude of ways, including a notable interaction with immune checkpoint inhibitor (ICI) cancer therapy.1 However, the specific mechanisms behind this interaction have remained cryptic, with some studies coming to contradictory conclusions regarding the effects exerted by the same bacterial species.2,3 Now, Dr. Rachel Newsome of the Jobin Lab at the University of Florida, in partnership with Isolation Bio, offers insights into how gut bacteria are influencing immunotherapy response.
As presented in a recent webinar, Dr. Newsome utilized the Prospector® microbial discovery platform to rapidly isolate bacterial strains from the gut microbiome, enabling the lab to screen for and identify specific bacterial metabolites that boost ICI response and may one day make ICI effective for more cancer patients.
The gut microbiome’s effect on immune checkpoint inhibition therapy
ICI functions by blocking inhibitory checkpoint receptors that allow cancer cells to evade immune response, thus “taking the foot off the break” and permitting an immune response to proceed against cancer cells. Efficacy is unfortunately low, with only 20% of patients exhibiting a positive therapeutic response to ICI treatment. However, an intriguing correlation has emerged between gut microbes and ICI response, with gut microbiota compositions enriched with Bacteroides species observed in responders compared to non-responders.1
In fact, when fecal samples from human ICI-responders and non-responders are transplanted into non-small cell lung cancer (NSCLC) mouse models, the resulting ICI response phenotypes in the mouse recipients mirror those of the human donor.4 This demonstrates that features inherent to the composition of the gut microbiome confer the responsive phenotype for this class of cancer immunotherapy.
Automated microbial isolation with the Prospector® identified the relevant bacteria
Dr. Newsome isolated bacteria from the feces of mice transplanted with responder gut microbiota using the 6000+ nanowell Prospector® array. The platform was then used to automatically pick and transfer isolates into 96-well plates for scaled culture. Microbial cultures were characterized by MALDI-TOF to identify genus and species, with 183 Bacteroides isolates identified, and 18 unique species represented (Figure 1).
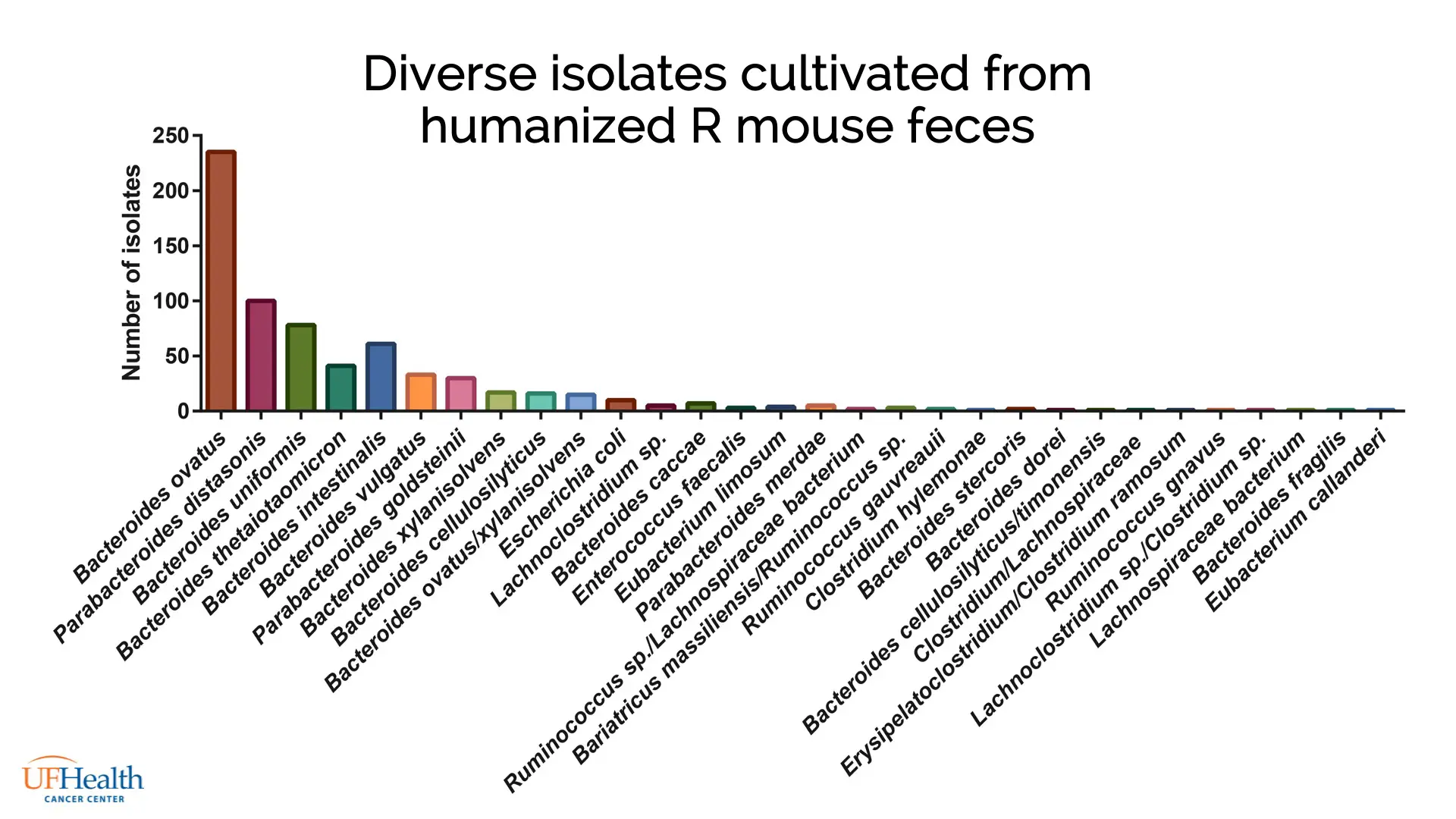
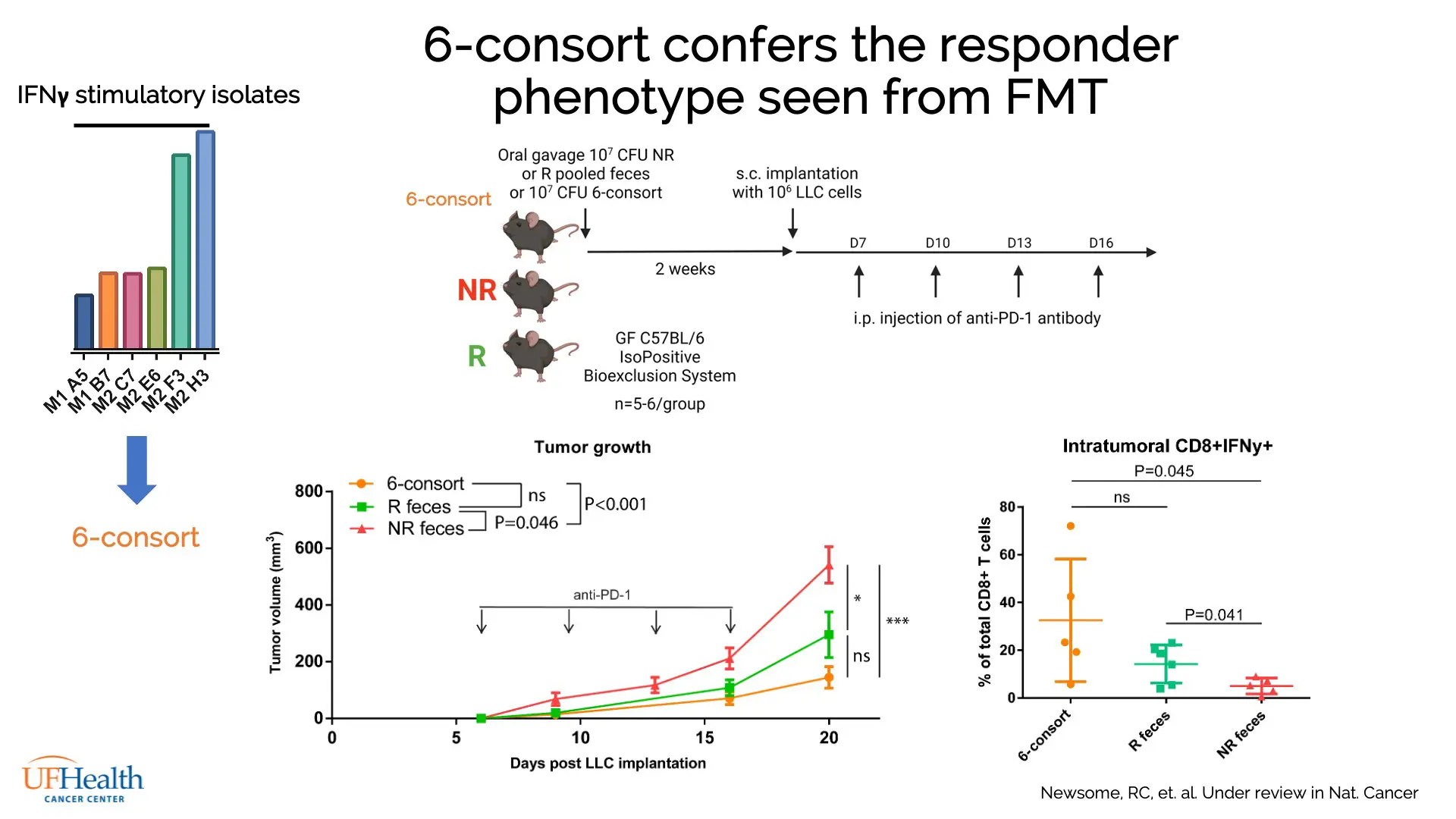
Supernatant from these cultures was applied to naïve CD8+ cytotoxic T cells cultured from wild-type mice, which were then examined for pro-inflammatory IFNγ production by flow cytometry. Supernatant from 6 species were shown to stimulate IFNγ, indicating their potential to affect ICI therapy.
Characterization and development of a promising new anti-tumor small molecule
When transplanted into mice, combined cultures from these 6 isolates, collectively termed the “6-consort” gut biota, conveyed the same ICI-responsive phenotype as fecal transplant from human ICI responders (Figure 2). Further, supernatant from 6-consort cultures, with molecules over 3kDa filtered out, was found to stimulate IFNγ in CD8+ T cells, showing the active agent to be a secreted small molecule.
With the active agent thus narrowed down, bioassay-guided fractionation of supernatant and another round of testing on T cells were used to identify a single IFNγ-stimulating molecule: cis-Bac429. This small molecule has now been synthesized and, when paired with ICI therapy, has been found effective at reducing tumor volume in an ICI-resistant NSCLC mouse model. Research is ongoing to determine its mechanism, bioactivity, and dose response.
Microbial-based products may improve cancer outcomes
The breakthrough discovery of cis-Bac429 was made possible with the use of the Prospector® platform. The Prospector® enables rapid isolation and cultivation of thousands of bacterial strains in parallel from complex microbiome samples. High-throughput processing accelerates the phenotyping of individual microbial species, the discovery of microbial attributes with an impact on human health, as well as the discovery of new therapeutic drug candidates.
Watch the webinar, “Small Microbes Make a Big Impact in Cancer Immunotherapy”, and hear Dr. Newsome present these findings in detail and discuss the promise of cis-Bac429 to improve outcomes for cancer patients.
References
- Vetizou, M. & Trinchieri, G. Anti-PD1 in the wonder-gut-land. Cell Res 28, 263–264 (2018).
- Routy, B. et al. Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science 359, 91–97 (2018).
- Lee, S.-H. et al. Bifidobacterium bifidum strains synergize with immune checkpoint inhibitors to reduce tumour burden in mice. Nat Microbiol 6, 277–288 (2021).
- Gharaibeh, R. Z. & Jobin, C. Microbiota and cancer immunotherapy: in search of microbial signals. Gut 68, 385–388 (2019).