Study of the switchgrass root microbiome is accelerated with the Prospector®
What if we could leverage the power of microbes to create a more sustainable energy source?
Switchgrass may one day provide enough biomass to enhance production of biofuel, a
renewable energy source. Studies are under way to evaluate whether bacterial species within the root microbiome can be manipulated to drive biomass production.
This task is not easy—the root microbiome consists of thousands of bacterial species and to identify and characterize them, each species must be isolated. Traditional Petri dish microbial culture methods are too slow and labor-intensive to rapidly generate large and highly diverse microbial isolate libraries for characterization. An automated isolation and cultivation system, however, can accelerate library generation as evidenced by research into the root microbiome of switchgrass.
Digging Into the Switchgrass Microbiome
Plants can alter the makeup of their own root microbiome by releasing compounds that either promote or hinder the growth of certain bacterial species within the roots or surrounding soil. Certain species of bacteria live symbiotically with plants, metabolizing plant compounds and proliferating. Bacteria subsequently convert soil nutrients into forms usable by the plant. Manipulating the bacterial populations in the root microbiome thus presents an opportunity to increase plant biomass for biofuel production. Altering the microbiome, rather than using more water and fertilizer, offers a sustainable alternative to increase plant biomass yield. Researchers at the University of Texas Austin, led by Dr. Thomas Juenger, are studying how the bacterial species and microbial profiles in the roots of switchgrass correlate with desirable plant qualities like higher biomass and drought tolerance. The team aims to uncover whether altering the switchgrass genome or root microbiome could enhance biomass yield.Slow Assembly of a Diverse Microbial Isolate Library
Revealing correlations between switchgrass growth and the activities of bacterial species in its root microbiome begins with isolating and cultivating a highly diverse microbial isolate library from the soil and roots of the plant. Once isolated, each species can be genetically and functionally characterized to determine its role in influencing plant growth. Traditional Petri dish culture methods are too labor-intensive and time consuming for isolating the thousands of microbial species that make up the switchgrass root microbiome. In addition, it is common for fast growing species to overtake slow-growing and rare species in Petri dishes, making it especially difficult to accurately capture microbial diversity. The Juenger lab spent 2 years using Petri dish culture methods with little return, generating a small library with low diversity, repeatedly isolating the same fast-growing species, and isolating species that were not of particular significance in the switchgrass microbiome according to historical sequencing data. To accelerate their research and improve the diversity of their isolate library, Joseph Edwards, Ph.D., of the Juenger lab implemented a significant change to their isolation and cultivation method.Switching Microbial Isolation and Cultivation Methods
Dr. Edwards enlisted the Prospector® to expedite their microbial isolation and cultivation workflows and increase the diversity of species obtained from switchgrass soil and root samples. The Prospector® uses automation and high throughput cultivation array technology to accelerate isolation and cultivation of live-cell workflows. Each array contains 6000+ nanoscale micro-wells, with each well capable of capturing a single bacterium and enabling the simultaneous growth of thousands of pure microcolonies. The physical separation of microcolonies enhances the ability to isolate slow-growing and rare species often lost in Petri dish culture due to competition with fast growing strains. Multiple nanoarrays can be used with multiple media and environmental conditions to capture greater microbial diversity. The Prospector® uses imaging to measure microbial growth within each nanowell and automates the picking and transfer of isolates into multi-well plates for characterization studies.Capturing Greater Microbial Diversity
The Juenger lab integrated the Prospector® into their isolation and cultivation workflow. After preparing just 2 soil microbiome samples and loading 4 nanoarrays on the Prospector®, 191 recultured isolates were selected for amplification with 16S rRNA primers and the amplicons were sequenced by the Sanger method. Analysis showed 98 individual species comprised of 48 genera. This level of diversity was significantly higher than what had been accomplished in the Juenger lab in over 2 years using Petri dish isolation and cultivation. Petri dish cultures had yielded repeated isolation of Bacillus and Pseudomonas species, yet these species are only minor constituents of the switchgrass root microbiome. The species isolated using the Prospector® correlated well with previous data on bacterial strains typically found in the soil surrounding switchgrass roots (Figure 1). Further study indicated that 27 of the 98 species showed sequence similarity to microbes previously linked to specific switchgrass phenotypes, underscoring the relevance of the strains isolated by the Prospector®. Additional data from this work can be found here.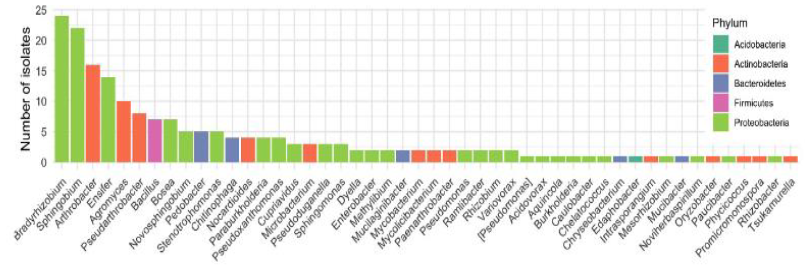
Having the isolated strains, the Juenger lab can now conduct further studies to determine how switchgrass genetic variations correlate with the growth of specific microbial strains in the root microbiome. Questions regarding which plant genes are influencing microbial growth and which microbial combinations can enhance plant growth, may now be answerable.
Conclusion
The Prospector® enabled the Juenger lab to rapidly build a diverse microbial library from the soil and roots of switchgrass plants, something that had not be achieved in over 2 years of Petri dish culture. With live isolates, the lab can now conduct additional characterization studies of each microbial species to determine the microbe-host relationship. This information may one day be leveraged to enhance switchgrass biomass and biofuel production.
Learn how the Prospector® makes it possible to rapidly assemble diverse microbial libraries and fuel microbial discovery.
Resources
Isolation Bio Case Study:
Use of the Prospector® System to Rapidly Generate Diverse Isolate Libraries for Correlation Studies of Switchgrass Phenotypes with the Root Microbiome